CRISPR Platforms for Stem Cell Biology
CRISPR-based functional genomics has transformed cell lines and cancer models, but it remains largely inaccessible in stem cell models of human development. A major obstacle is the pervasive silencing of CRISPR machinery during differentiation, which renders most transgene-based tools ineffective in long-term contexts like 3D-organoids. This bottleneck has severely limited our ability to dissect how key developmental regulators control cell fate, maturation, and tissue architecture.
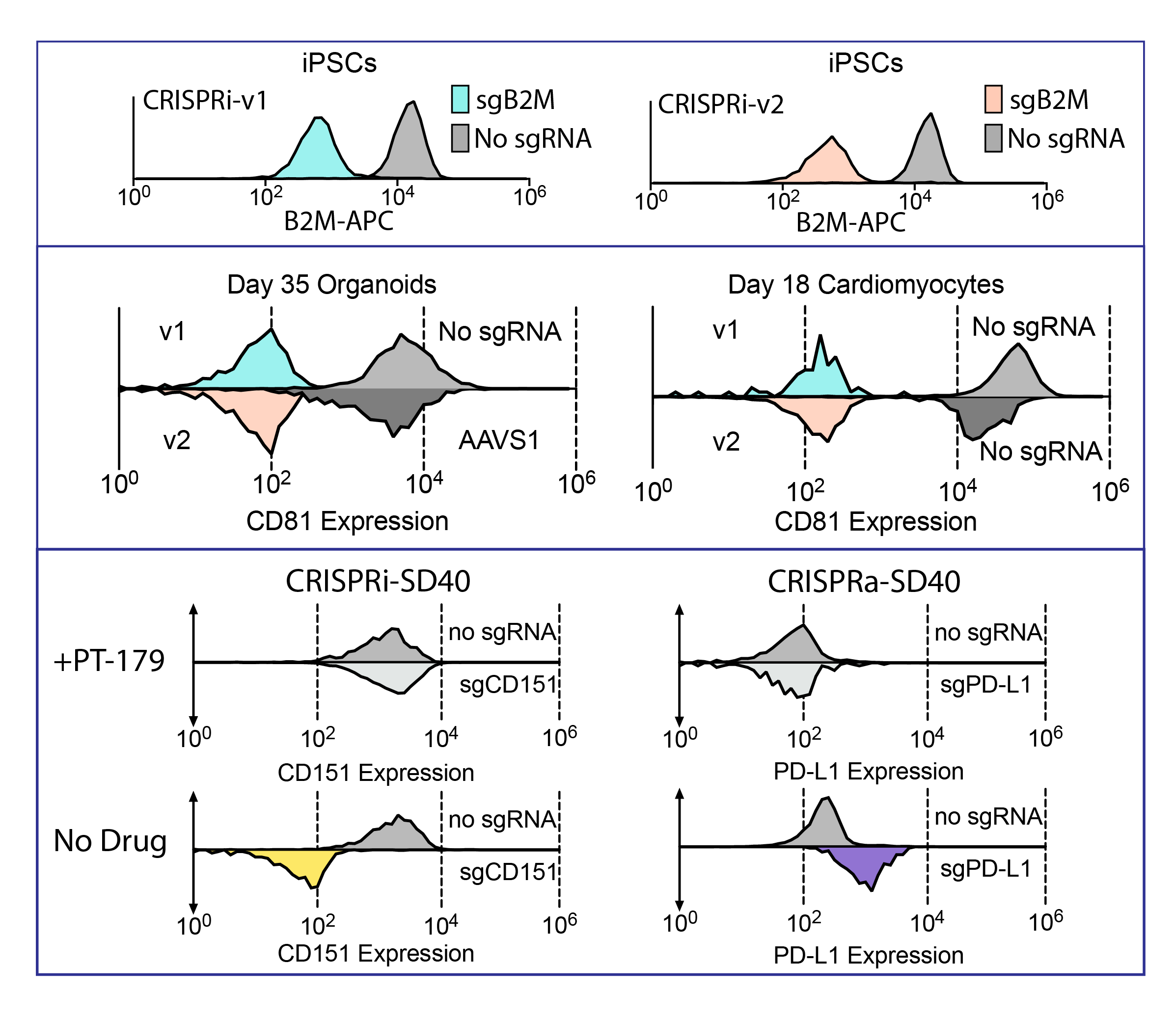
Our lab has developed next-generation CRISPRi and CRISPRa platforms that overcome this barrier. Our latest tools achieve robust expression of dCas9 effectors across all three germ layers, including in 35-day cortical organoids, 18-day cardiomyocytes, and hepatocytes. We routinely observe ~99% repression of easily observable surface markers such as CD81 or B2M (Fig 1), enabling true genome-wide perturbation screens in differentiated human tissues for the first time.
To add temporal precision, we engineered a degron-based CRISPR system using the SD40 tag, which allows rapid ON/OFF switching via a small-molecule inducer. This inducible Cas9 exhibits zero leakiness and full reversibility, enabling dynamic, time-resolved perturbation experiments throughout human stem cell differentiation.
We view this platform as a foundation for a new era of stem-cell–based functional genomics—one that enables precise, large-scale dissection of the gene networks that govern lineage specification, functional maturation, and tissue integration. By combining genome-scale perturbation with temporal control, we can now ask questions such as:
- Which genes refine the trajectory of differentiation toward specific, therapeutically relevant cell types? For example, pancreatic beta cell differentiation from iPSCs remains inefficient; systematic loss-of-function and overexpression screens could identify regulators that enhance lineage fidelity and suppress off-target fates.
- Which pathways promote the functional maturation of in vitro–derived cell types? Many systems, including cortical organoids and cardiomyocytes, stall in a fetal-like state. CRISPR-based screening offers a way to discover transcriptional or epigenetic regulators that drive late-stage maturation and functional competence.
- Which gene perturbations enhance the survival, engraftment, or integration of transplanted cells in vivo? Dopaminergic neuron transplants for Parkinson’s disease, now in clinical trials, highlight both the promise and the challenges of cell therapy. Genetic screens could uncover factors that improve long-term graft viability and function in host tissue.
These questions span tissues and disease areas far beyond what we can pursue alone. While our focus is on the nervous system, the tools are broadly applicable. We're eager to collaborate—please get in touch if you're interested in deploying them in your own stem cell system.
A Functional Genomics Map of Human Brain Evolution
The three‑fold expansion of the human cerebral cortex is perhaps the most defining feature of our evolution. This dramatic growth, which unfolded over just a few million years, enabled the emergence of language, abstract reasoning, and complex culture—but we still don’t know which genes or pathways actually drove it.
Our lab tackles this question head‑on by combining cerebral organoid models, genome‑wide CRISPR perturbation, and cross‑species comparisons. Using human and chimpanzee iPSCs, we grow 3D cortical organoids side‑by‑side and perform pooled CRISPRi/a screens to identify genes that regulate progenitor self‑renewal, neurogenesis, and differentiation dynamics. With a custom‑designed guide library that perfectly matches both genomes, we measure how each perturbation shifts the balance of cell types during development—then ask whether those effects are conserved or species‑specific.
Key questions that we seek to answer include:
- How is the balance between progenitor self-renewal and neurogenic commitment tuned across primate species? Which signaling pathways, epigenetic regulators, or metabolic pathways play a key role?
- Can outgroup comparisons across the great apes reveal unique molecular changes on the hominin branch of evolution?
- Where do evolutionary innovations intersect with loci implicated in neurodevelopmental disorders, and does human cortical expansion lead to unique vulnerabilities?